143 lines
No EOL
6.4 KiB
Text
143 lines
No EOL
6.4 KiB
Text
---
|
|
title: "General Feature index (GFidx)"
|
|
subtitle: "Space-efficient and fast index for querying large General Feature Format (GFF) files"
|
|
author: ["<redacted>"]
|
|
date: "2024-08-01"
|
|
format:
|
|
revealjs:
|
|
transition: fade
|
|
theme: sakura.scss
|
|
slideNumber: true
|
|
embed-resources: true
|
|
csl: american-chemical-society.csl
|
|
bibliography: presentation.bib
|
|
---
|
|
|
|
## Introduction {.smaller}
|
|
|
|
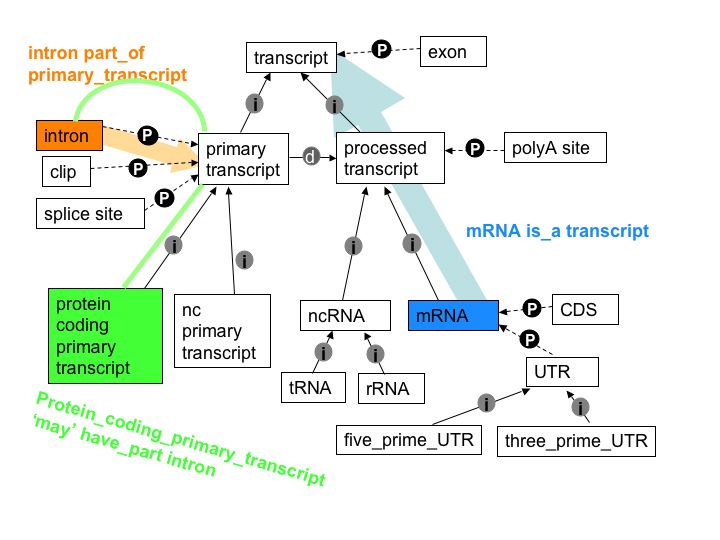
- General Feature Format (GFF) files are used to store genomic features and annotations. The GFF3 format is a widely used standard for representing genomic feature hierarchies.
|
|
|
|
:::: {.columns}
|
|
|
|
::: {.column width="50%"}
|
|
|
|
:::
|
|
|
|
::: {.column width="50%"}
|
|
Each feature in a GFF file is represented as a line of tab-separated fields. The fields include:
|
|
|
|
- The range and location of the feature
|
|
- The Sequence Ontology (SO) [@noauthor_httpwwwsequenceontologyorg_nodate] term of the feature
|
|
- The Sequence Ontology (SO) parent of the feature
|
|
- Other attributes such as gene name, gene type, aliases, etc.
|
|
:::
|
|
::::
|
|
|
|
|
|
## Motivation {.smaller}
|
|
|
|
- GFF files can be very large, making it difficult to query them efficiently:
|
|
- In the Human GENCODE GFF3 file, there are 3.7 million features. A lot of genes have more than 400 features associated with them.
|
|
|
|
:::: {.columns}
|
|
|
|
::: {.column width="50%"}
|
|
- Often only a subset of the features are needed for analysis.
|
|
- Inefficient to load the entire file into memory.
|
|
- In a cluster environment, the whole file may be split across multiple nodes
|
|
- Looking up a feature by ID or attribute requires a linear scan of the file, which is time and resource consuming.
|
|
:::
|
|
::: {.column width="50%"}
|
|
- Example pipelines and operations include:
|
|
- Gene-centric analysis: large number of ID -> feature queries
|
|
- RNA alternative splicing analysis: large number of exon -> gene queries
|
|
- Genome browser: range to feature queries
|
|
- General queries: Ancestors, descendants and overlaps of a feature
|
|
:::
|
|
|
|
::::
|
|
|
|
## Existing Solutions
|
|
|
|
- Heuristic-based solutions:
|
|
- Assume that features are ordered in a way that related features are close to each other.
|
|
- No guarantee that features are ordered in this way
|
|
- Only solves the problem for looking up by relation
|
|
- Relational databases:
|
|
- Load the GFF file into a database
|
|
- Requires predefined schema for the attributes
|
|
- Querying by range or name/ID prefix is still slow
|
|
|
|
## GFidx - General Feature index {.smaller}
|
|
|
|

|
|
|
|
- A range index is first built to save the approximate location of each feature in the GFF file. This allows further indexing to only save a reference to the feature in the GFF file instead of the whole feature.
|
|
- Two additional types of indices are built:
|
|
- Attribute indices: for looking up features by ID or attribute prefix
|
|
- Relation tree: for looking up features by parent-child relationships
|
|
|
|
## Methodology - Lookup by range {.smaller}
|
|
|
|
:::: {.columns}
|
|
::: {.column width="40%"}
|
|
- For each sequence, split the range into non-overlapping intervals and record the location of the first and last feature that starts in each interval in the GFF file.
|
|
- Split the intervals recursively until the number of features in each interval is small enough, forming a tree structure.
|
|
- Some extra data may be returned but by controlling the granularity of the tree, a balance between the amount of unnecessary data and the size of the index can be achieved.
|
|
:::
|
|
::: {.column width="60%"}
|
|
{style="width: 100%"}
|
|
:::
|
|
::::
|
|
|
|
## Methodology - Lookup by attribute {.smaller}
|
|
|
|
- Instead of recording the IDs and attributes by feature, we sort them into a Trie structure, and only record the location of the corresponding feature in the GFF file.
|
|
- Fast lookup by ID or attribute prefix.
|
|
- Space-efficient: the long ENSEMBL gene IDs are stored only once.
|
|
- Fast to build: Only one pass through the GFF file is needed.
|
|
|
|
{style="width: 100%"}
|
|
|
|
## Methodology - Lookup by relation {.smaller}
|
|
|
|
- Parent-child relationships are stored in a tree structure.
|
|
- Each node contains the range of the feature and a list of children.
|
|
- The tree is built by iterating through the GFF file and adding each feature to the corresponding parent node in the tree.
|
|
- Since GFF3 requires all relations to be in the Sequence Ontology structure, the tree structure can be pre-built and reused for different queries.
|
|
|
|
{style="width: 100%"}
|
|
|
|
## Results {.smaller}
|
|
|
|
- A full index of the Human GENCODE [@frankish_gencode_2019] GFF3 file (1.62 GB decompressed) can be built using 3 processors in less than 20 seconds.
|
|
- All queries are done in less than 0.2 seconds. Demo queries include:
|
|
- All features of CDCA8 (Cell Division Cycle Associated 8) gene - 34 features, 600 $\mu$s, 272 KB downloaded.
|
|
- All genes in the SLC35 family (Nucleotide Sugar Transporter Family) - 2,926 features, 128 ms, 22 MB downloaded.
|
|
- All genes in chr3 from 650,000 to 1,500,000 bp - 243 features, 1.70 ms, 120 KB downloaded.
|
|
- More effort is needed to optimize the space efficiency of the index:
|
|
- Simply using an off-the-shelf binary format, the uncompressed index containing all relavant attributes is 268 MB.
|
|
- Most of the space is used by the relation index, nearly 100 MB.
|
|
|
|
## Conclusion
|
|
|
|
- GFidx has a potential to be a useful tool for both in-memory and over-the-network querying of large GFF files in certain pipelines and applications.
|
|
- Web-based services could benefit from GFidx by only downloading the relevant parts of the GFF file.
|
|
- Further space optimization and parallelization could be done especially on the trie building and relation tree building steps.
|
|
|
|
## Future Work {.smaller}
|
|
|
|
- More research on common queries and operations on GFF files.
|
|
- More flexibility on the granularity of indexes to save time and space.
|
|
- More efficient encoding of feature locations.
|
|
- Compression of the index using BGZF (used in BAM [@noauthor_bam_nodate] files).
|
|
- Parallelization of the trie and tree building.
|
|
- A WebAssembly build for use in web applications such as IGV Genome Browser: only download the file range that is currently being viewed.
|
|
- Support for features with multiple parents (allowed in GFF3 but rarely used).
|
|
- Test suites for real-world GFF3 files.
|
|
- Integration with existing bioinformatics tools and pipelines.
|
|
|
|
## References
|
|
|
|
::: {#refs}
|
|
::: |